bio gff: convert to GFF
Building a nicer gene model
bio creates more meaningful and nicer GFF visualizations. Get data for chromosome 2L for Drosophila melanogaster (fruit-fly)
bio fetch NT_033779 > chrom2L.gbConvert it to gff:
bio gff chrom2L.gb > chrom2L.gffGFF created with bio
Here is a region from the GFF file created with the code above as visualized in IGV:
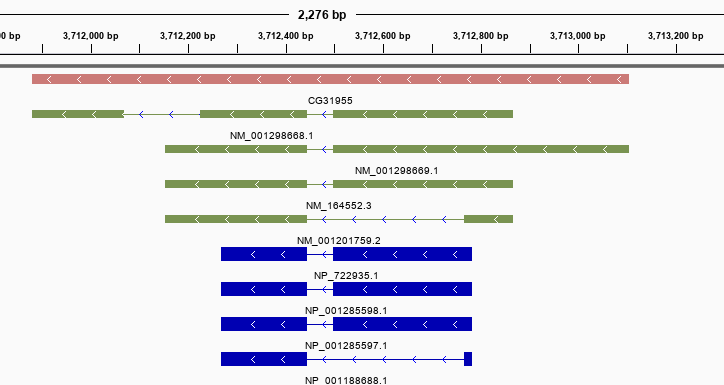
- Exons will have
transcript_idandgene_idattributes set. - CDS features have
protein_idandgene_idattributes set.
Get a dataset
Get SARS-COV-2 data:
bio fetch NC_045512 MN996532 > genomes.gbConvert all features to GFF:
cat genomes.gb | bio gff | head -5Convert to GFF only the features with type CDS
cat genomes.gb | bio gff --type gene,CDS | head -5 Convert to GFF only the features tagged with gene S
bio gff --gene S genomes.gb | head -5